Welcome
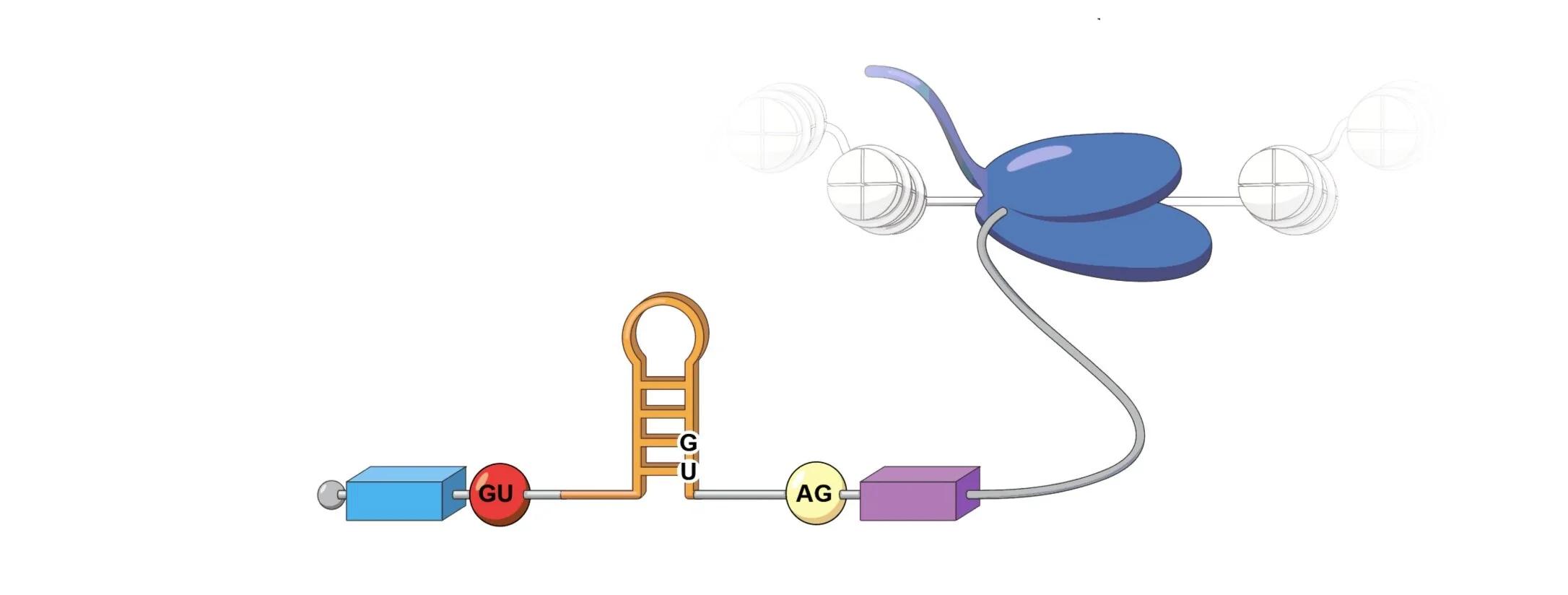
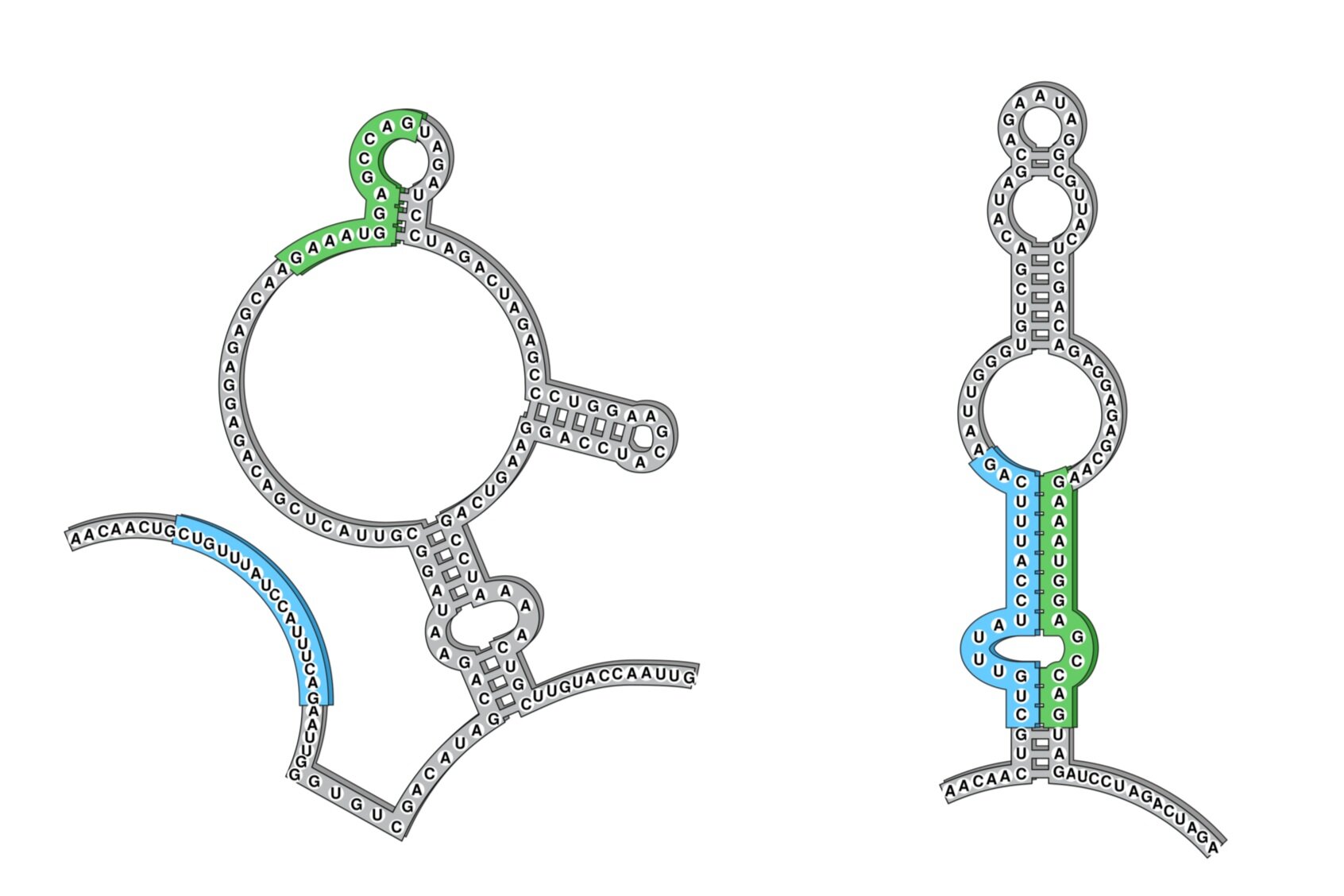
Our lab is interested in alternative RNA structures and the myriad of roles they have in both viral and human biology. In particular, we study how RNA folding informs alternative splicing and how mis-folding can lead to disease. Our goal is to understand gene expression regulation in physiology and disease through the lens of RNA.
We developed DMS-MaPseq (dimethyl sulfate mutational profiling with sequencing) and DREEM (Detection-of-RNA-folding-Ensembles-using-Expectation-Maximization) algorithm to distinguish multiple RNA conformations formed by the same underlying sequence in vivo at single nucleotide resolution at a high-throughput scale.
Alternative RNA Structures
DMS
Mapseq